Google’s DeepMind Makes a Quantum Leap in Solving the Protein Folding Problem
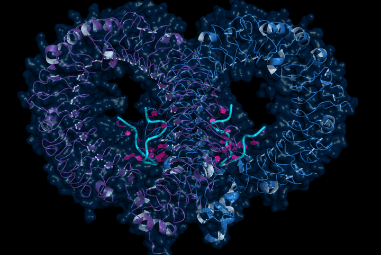
Artificial Intelligence has made something of a breakthrough in the prediction of protein structures. The results came about as part of the 14th Critical Assessment of Structure Prediction, a friendly contest and conference organized by the Protein Structure Prediction Center with sponsorship from the US NIH’s National Institute of General Medical Sciences (NIH/NIGMS). The challengers in the competition are research teams that have developed software methods for determining a protein’s physical structure from the sequence of amino acids composing it. While performance has improved over the 14 biennial iterations of the CASP challenge, the mark set this year by the winning competitor was so far in advance of the winning performance in 2018, that many thought it would take a decade to achieve such a level. The winning software in both 2018 and 2020 was AlphaFold, a product of DeepMind Technologies, an artificial intelligence firm acquired by Google in 2014.
While the chemical identity of a protein is uniquely specified by the string of amino acids that compose it, much a protein’s functional properties derive from its shape, and this three-dimensional structure is not immediately obvious from building up the chain of amino acids. Understanding the structure helps provide information on how a protein performs its function, and further provides clues to how that function can be promoted or retarded through therapeutic interventions that can target certain parts of the structure. Although it might be easy to link together two or three amino acids and fully develop the molecular structure based on the electric charges of the component atoms, typical human proteins are composed of hundreds of amino acids, with the median number over 400. At this scale, the self-interactions of all of the links in the chain as it folds together is tremendously complicated. Traditionally, structures have been determined by actually studying the physical proteins, rather than attempting to predict their structures through their composition. The primary methods for this have been x-ray crystallography, based on x-ray diffraction (XRD), nuclear magnetic resonance (NMR), and the newer technique of cryo-electron microscopy, which combines electron microscopy with cryogenically frozen samples. All of these are roughly complementary techniques that have different strengths and weaknesses in terms of ease of sample preparation, speed of experiment, and quality of results.
To return to AlphaFold and the CASP challenge, as DeepMind recounts their success, the 2018 version of the software achieved a modelling accuracy approaching 60% when compared to the results of structure determination by traditional methods. In 2020, this accuracy improved to nearly 90%. In some cases, the software-predicted structures are indistinguishable from the ‘right answer’ within the uncertainties inherent in the traditional methods. From the perspective of the analytical instrumentation industry, the obvious question is: what does this mean for XRD, NMR, and electron microscopy? Have these instruments and the scientists who use them been surpassed and supplanted by AI, like chess grandmasters and Jeopardy contestants? Fortunately, the answer is not yet. Despite this significant leap, the AI solutions have not yet been perfected, and traditional techniques will still be required to provide data for the AI to learn from, and to verify or improve the software solutions. However, it is very likely that within a shorter time than might have been expected, the bulk of the heavy lifting will be done in silico. For more information about the instrumentation markets involved in protein structure determination and hundreds of other applications, SDi’s 2020 Global Assessment Report provides valuable insights and market data on more than 80 laboratory technologies.